eDNA as a Population Genetics Tool
Environmental DNA (eDNA) has the potential to provide a more comprehensive perspective on community-wide population structure by increasing species sampled in meta-analyses to hundreds. Previous studies in marine ecosystems have demonstrated that eDNA can reliably detect haplotypes. However, a key challenge with eDNA as a population genetics tool is determining haplotype frequencies from environmental sequence pools from many individuals. For organisms with high gene flow, fine-scale population structure is often limited to differences in haplotype frequencies rather than haplotype presence and absence. Therefore, the scale at which population structure can be detected with eDNA methods is unknown.
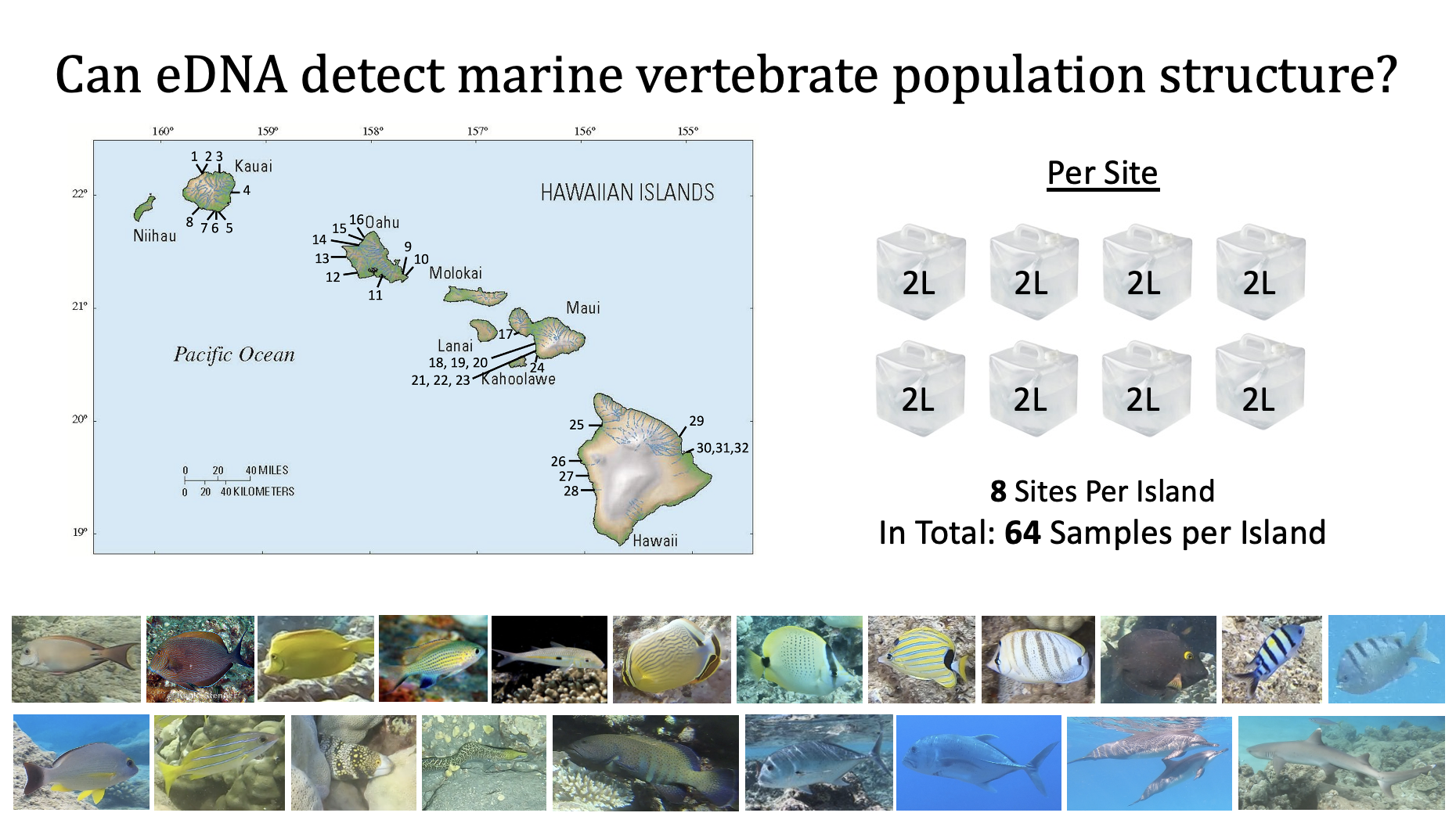
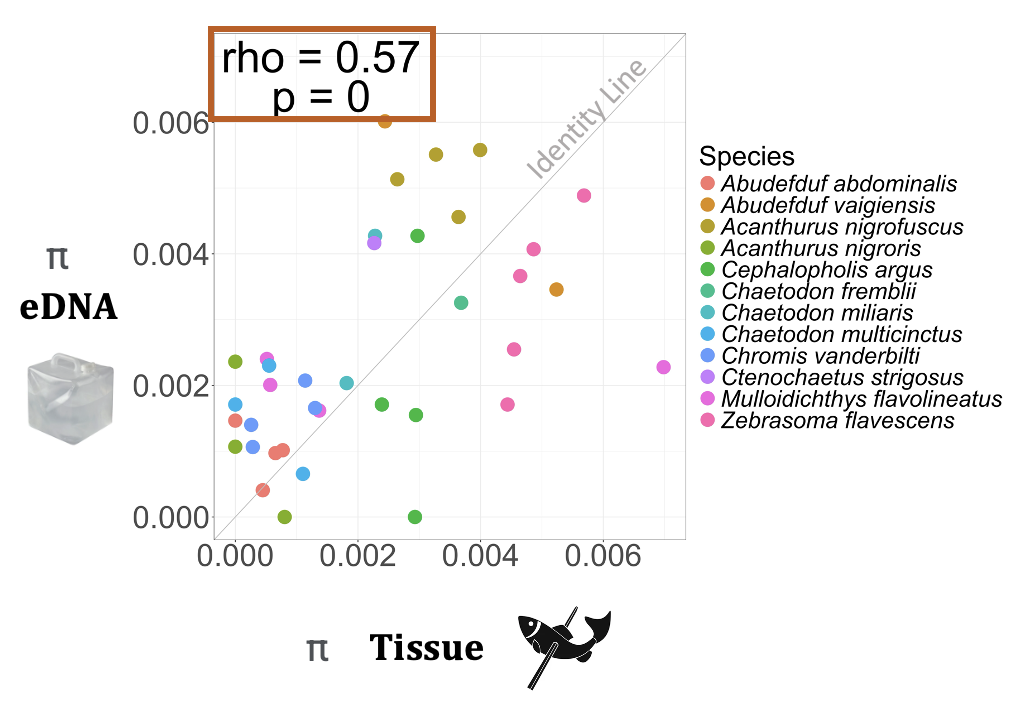
For my PhD thesis I am investigating whether eDNA methods can accurately describe mtDNA sequence diversity and detect population structure in Hawaiian marine fish communities previously studied using traditional, individual-based (i.e., tissue-based) methods. Among 18 species, I found significant correlations in mtDNA sequence diversity metrics (i.e., nucleotide and haplotype diversity) between eDNA and tissue-based methods, even at low sample sizes. While eDNA methods detected similar haplotypes at similar frequencies as tissue-based methods, eDNA detected a higher diversity of haplotypes for most species.
However, using eDNA as a tool for analyzing population structure showed that only at larger sample sizes were eDNA and tissue-based population structure metrics (i.e., ΦST) weakly but significantly correlated. eDNA detected more significant population structure than tissue-based results. eDNA methods may have greater power to detect population subdivisions because eDNA methods often capture DNA from larger numbers of individuals.